This is part II of our series of posts discussing Crail's raw storage performance. This part is about Crail's NVMe storage tier, a low-latency flash storage backend for Crail completely based on user-level storage access.
Hardware Configuration
The specific cluster configuration used for the experiments in this blog:
- Cluster
- 8 node OpenPower cluster
- Node configuration
- CPU: 2x OpenPOWER Power8 10-core @2.9Ghz
- DRAM: 512GB DDR4
- 4x 512 GB Samsung 960Pro NVMe SSDs (512Byte sector size, no metadata)
- Network: 1x100Gbit/s Mellanox ConnectX-4 IB
- Software
- RedHat 7.3 with Linux kernel version 3.10
- Crail 1.0, internal version 2843
- SPDK git commit 5109f56ea5e85b99207556c4ff1d48aa638e7ceb with patches for POWER support
- DPDK git commit bb7927fd2179d7482de58d87352ecc50c69da427
The Crail NVMf Storage Tier
Crail is a framework that allows arbitrary storage backends to be added by implementing the Crail storage interface. A storage backend manages the point-to-point data transfers on a per block granularity between a Crail client and a set of storage servers. The Crail storage interface essentially consists of three virtual functions, which simplified look like this:
//Server-side interface: donate storage resources to Crail
StorageResource allocateResource();
//Client-side interface: read/write remote/local storage resources
writeBlock(BlockInfo, ByteBuffer);
readBlock(BlockInfo, ByteBuffer);
A specific implementation of this interface provides an efficient mapping of Crail storage operations to the actual storage and network hardware the backend is exporting. Crail comes with two native storage backends, an RDMA-based DRAM backend and an RDMA-based NVMe backend, but other storage backends are available as well (e.g., Netty) and we plan to provide more custom backends in the future as new storage and network technologies are emerging.
The Crail NVMf storage backend we evaluate in this blog provides user-level access to local and remote flash through the NVMe over Fabrics protocol. Crail NVMf is implemented using DiSNI, a user-level network and storage interface for Java offering both RDMA and NVMf APIs. DiSNI itself is based on SPDK for its NVMf APIs.
The server side of the NVMf backend is designed in a way that each server process manages exactly one NVMe drive. On hosts with multiple NVMe drives one may start several Crail NVMf servers. A server is setting up an NVMf target through DiSNI and implements the allocateResource() storage interface by allocating storage regions from the NVMe drive (basically splits up the NVMe namespace into smaller segments). The Crail storage runtime makes information about storage regions available to the Crail namenode, from where regions are further broken down into smaller units called blocks that make up files in Crail.
The Crail client runtime invokes the NVMf client interface during file read/write operations for all data transfers on NVMf blocks. Using the block information provided by the namenode, the NVMf storage client implementation is able to connect to the appropriate NVMf target and perform the data operations using DiSNI's NVMf API.
One downside of the NVMe interface is that byte level access is prohibited. Instead data operations have to be issued for entire drive sectors which are typically 512Byte or 4KB large (we used 512Byte sector size in all the experiments shown in this blog). As we wanted to use the standard NVMf protocol (and Crail has a client driven philosophy) we needed to implement byte level access at the client side. For reads this can be achieved in a straight forward way by reading the whole sector and copying out the requested part. For writes that modify a certain subrange of a sector that has already been written before we need to do a read modify write operation.
Performance comparison to native SPDK NVMf
We perform latency and throughput measurement of our Crail NVMf storage tier against a native SPDK NVMf benchmark to determine how much overhead our implementation adds. The first plot shows random read latency on a single 512GB Samsung 960Pro accessed remotely through SPDK. For Crail we also show the time it takes to perform a metadata operations. You can run the Crail benchmark from the command line like this:
./bin/crail iobench -t readRandom -b false -s <size> -k <iterations> -w 32 -f /tmp.dat
and SPDK:
./perf -q 1 -s <size> -w randread -r 'trtype:RDMA adrfam:IPv4 traddr:<ip> trsvcid:<port>' -t <time in seconds>
The main take away from this plot is that the time it takes to perform a random read operation on a NVMe-backed file in Crail takes only about 7 microseconds more time than fetching the same amount of data over a point-to-point SPDK connection. This is impressive because it shows that using Crail a bunch of NVMe drives can be turned into a fully distributed storage space at almost no extra cost. The 7 microseconds are due to Crail having to look up the specific NVMe storage node that holdes the data -- an operation which requires one extra network roundtrip (client to namenode). The experiment represents an extreme case where no metadata is cached at the client. In practice, file blocks are often accessed multiple times in which case the read latency is further reduced. Also note that unlike SPDK which is a native library, Crail delivers data directly into Java off-heap memory.

The second plot shows sequential read and write throughput with a transfer size of 64KB and 128 outstanding operations. The Crail throughput benchmark can be run like this:
./bin/crail iobench -t readAsync -s 65536 -k <iterations> -b 128 -w 32 -f /tmp.dat
and SPDK:
./perf -q 128 -s 65536 -w read -r 'trtype:RDMA adrfam:IPv4 traddr:<ip> trsvcid:<port>' -t <time in seconds>
For sequential operations in Crail, metadata fetching is inlined with data operations as described in the DRAM blog. This is possible as long as the data transfer has a lower latency than the metadata RPC, which is typically the case. As a consequence, our NVMf storage tier reaches the same throughput as the native SPDK benchmark (device limit).

Sequential Throughput
Let us look at the sequential read and write throughput for buffered and direct streams and compare them to a buffered Crail stream on DRAM. All benchmarks are single thread/client performed against 8 storage nodes with 4 drives each, cf. configuration above. In this benchmark we use 32 outstanding operations for the NVMf storage tier buffered stream experiments by using a buffer size of 16MB and a slice size of 512KB, cf. part I. The buffered stream reaches line speed at a transfer size of around 1KB and shows only slightly slower performance when compared to the DRAM tier buffered stream. However we are only using 2 outstanding operations with the DRAM tier to achieve these results. Basically for sizes smaller than 1KB the buffered stream is limited by the copy speed to fill the application buffer. The direct stream reaches line speed at around 128KB with 128 outstanding operations. Here no copy operation is performed for transfer size greater than 512Byte (sector size). The command to run the Crail buffered stream benchmark:
./bin/crail iobench -t read -s <size> -k <iterations> -w 32 -f /tmp.dat
The direct stream benchmark:
./bin/crail iobench -t readAsync -s <size> -k <iterations> -b 128 -w 32 -f /tmp.dat

Random Read Latency
Random read latency is limited by the flash technology and we currently see around 70us when performing sector size accesses to the device with the Crail NVMf backend. In comparison, remote DRAM latencies with Crail are about 7-8x faster. However, we believe that this will change in the near future with new technologies like PCM. Intel's Optane drives already can deliver random read latencies of around 10us. Considering that there is an overhead of around 10us to access a drive with Crail from anywhere in the cluster, using such a device would put random read latencies somewhere around 20us which is only half the performance of our DRAM tier.

Tiering DRAM - NVMf
In this paragraph we show how Crail can leverage flash memory when there is not sufficient DRAM available in the cluster to hold all the data. As described in the overview section, if you have multiple storage tiers deployed in Crail, e.g. the DRAM tier and the NVMf tier, Crail by default first uses up all available resources of the faster tier. Basically a remote resource of a faster tier (e.g. remote DRAM) is preferred over a slower local resource (e.g., local flash), motivated by the fast network. This is what we call horizontal tiering.
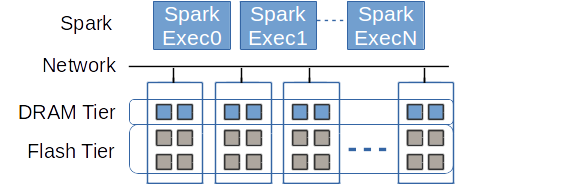
In the following 200G Terasort experiment we gradually limit the DRAM resources in Crail while adding more flash to the Crail NVMf storage tier. Note that here Crail is used for both input/output as well as shuffle data. The figure shows that by putting all the data in flash we only increase the sorting time by around 48% compared to the configuration where all the data resides in DRAM. Considering the cost of DRAM and the advances in technology described above we believe cheaper NVM storage can replace DRAM for most of the applications with only a minor performance decrease. Also, note that even with 100% of the data in NVMe, Spark/Crail is still faster than vanilla Spark with all the data in memory. The vanilla Spark experiment uses Alluxio for input/output and RamFS for the shuffle data.

To summarize, in this blog we have shown that the NVMf storage backend for Crail – due to its efficient user-level implementation – offers latencies and throughput very close to the hardware speed. The Crail NVMf storage tier can be used conveniently in combination with the Crail DRAM tier to either save cost or to handle situations where the available DRAM is not sufficient to store the working set of a data processing workload.
